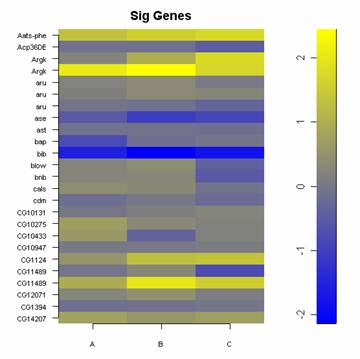
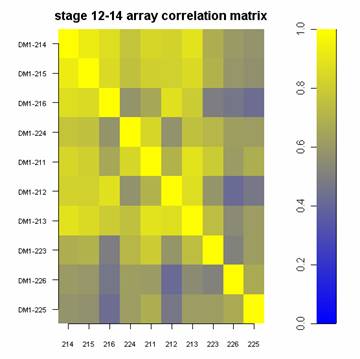
placed on several different DNA microarrays correlate with one another.
Solution: Use the function below by handing it a matrix of numbers. It will plot the matrix with a color scale based on the highest and lowest values in the matrix. Optional arguments are:
usage: myImagePlot(m) where m is a matrix of numbers
optional arguments: myImagePlot(m, xlabels, ylabels, zlim, title=c("my title"))

| 
|
| 1.) A panel of gene expression values. | 2.) A table of correlation values illustrating how identical samples placed on several different DNA microarrays correlate with one another. |
The images above were generated as follows:
myImagePlot(bm[1:25,1:3], yLabels=c(as.character(baylies[1:25,2])), title=c("Sig Genes"))
myImagePlot(cm, xLabels=c(targets$SlideNumber), title=c("stage 12-14 array correlation matrix"), zlim=c(0,1))
You can load the function below into your R session by copy and paste, or you can load it directly using the source command:
source("http://www.phaget4.org/R/myImagePlot.R")
| image plot function |
# ----- Define a function for plotting a matrix ----- #
myImagePlot <- function(x, ...){
min <- min(x)
max <- max(x)
yLabels <- rownames(x)
xLabels <- colnames(x)
title <-c()
# check for additional function arguments
if( length(list(...)) ){
Lst <- list(...)
if( !is.null(Lst$zlim) ){
min <- Lst$zlim[1]
max <- Lst$zlim[2]
}
if( !is.null(Lst$yLabels) ){
yLabels <- c(Lst$yLabels)
}
if( !is.null(Lst$xLabels) ){
xLabels <- c(Lst$xLabels)
}
if( !is.null(Lst$title) ){
title <- Lst$title
}
}
# check for null values
if( is.null(xLabels) ){
xLabels <- c(1:ncol(x))
}
if( is.null(yLabels) ){
yLabels <- c(1:nrow(x))
}
layout(matrix(data=c(1,2), nrow=1, ncol=2), widths=c(4,1), heights=c(1,1))
# Red and green range from 0 to 1 while Blue ranges from 1 to 0
ColorRamp <- rgb( seq(0,1,length=256), # Red
seq(0,1,length=256), # Green
seq(1,0,length=256)) # Blue
ColorLevels <- seq(min, max, length=length(ColorRamp))
# Reverse Y axis
reverse <- nrow(x) : 1
yLabels <- yLabels[reverse]
x <- x[reverse,]
# Data Map
par(mar = c(3,5,2.5,2))
image(1:length(xLabels), 1:length(yLabels), t(x), col=ColorRamp, xlab="",
ylab="", axes=FALSE, zlim=c(min,max))
if( !is.null(title) ){
title(main=title)
}
axis(BELOW<-1, at=1:length(xLabels), labels=xLabels, cex.axis=0.7)
axis(LEFT <-2, at=1:length(yLabels), labels=yLabels, las= HORIZONTAL<-1,
cex.axis=0.7)
# Color Scale
par(mar = c(3,2.5,2.5,2))
image(1, ColorLevels,
matrix(data=ColorLevels, ncol=length(ColorLevels),nrow=1),
col=ColorRamp,
xlab="",ylab="",
xaxt="n")
layout(1)
}
# ----- END plot function ----- #
|